Examples¶
Performing FFT on the raw data to get a view of the estimated depth of cell features¶
1# -*- coding: utf-8 -*-
2# Copyright 2018-2021 Nick Anthony, Backman Biophotonics Lab, Northwestern University
3#
4# This file is part of PWSpy.
5#
6# PWSpy is free software: you can redistribute it and/or modify
7# it under the terms of the GNU General Public License as published by
8# the Free Software Foundation, either version 3 of the License, or
9# (at your option) any later version.
10#
11# PWSpy is distributed in the hope that it will be useful,
12# but WITHOUT ANY WARRANTY; without even the implied warranty of
13# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
14# GNU General Public License for more details.
15#
16# You should have received a copy of the GNU General Public License
17# along with PWSpy. If not, see <https://www.gnu.org/licenses/>.
18
19"""
20Load the OPD from a previously saved analysis result and plot it using a special multi-dimensional plotting widget.
21
22@author: Nick Anthony
23"""
24
25import pwspy.dataTypes as pwsdt
26from mpl_qt_viz.visualizers import PlotNd
27from examples import PWSImagePath
28import matplotlib.pyplot as plt
29
30plt.ion() # Without this we will get a crash when trying to open the PlotNd window because a Qt application loop must be running.
31plt.figure()
32
33acquisition = pwsdt.Acquisition(PWSImagePath) # Get a reference to the top-level folder for the measurement.
34
35roiSpecs = acquisition.getRois() # Get a list of the (name, number, file format) of the available ROIs.
36print("ROIs:\n", roiSpecs)
37
38analysis = acquisition.pws.loadAnalysis(acquisition.pws.getAnalyses()[0]) # Load a reference to an analysis file.
39kCube = analysis.reflectance # Load the processed `reflectance` array from the analysis file.
40
41opd, opdValues = kCube.getOpd(useHannWindow=False, indexOpdStop=50) # Use FFT to transform the reflectance array to OPD
42
43# Scale the opdValues to give estimated depth instead of raw OPD. Factor of 2 because light is making a round-trip.
44ri = 1.37 # Estimated RI of livecell chromatin
45opdValues = opdValues / (2 * ri)
46
47plotWindow = PlotNd(opd, names=('y', 'x', 'depth'),
48 indices=(None, None, opdValues), title="Estimated Depth")
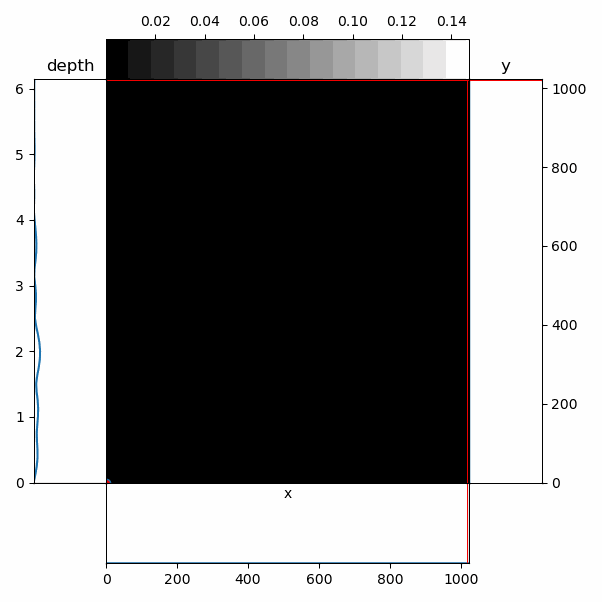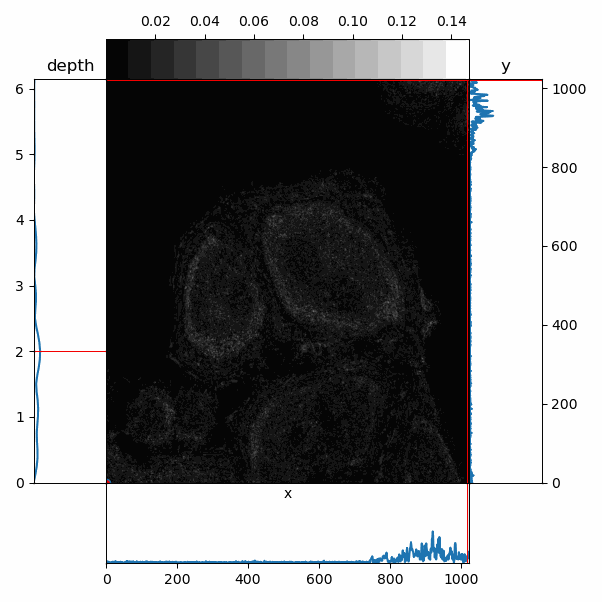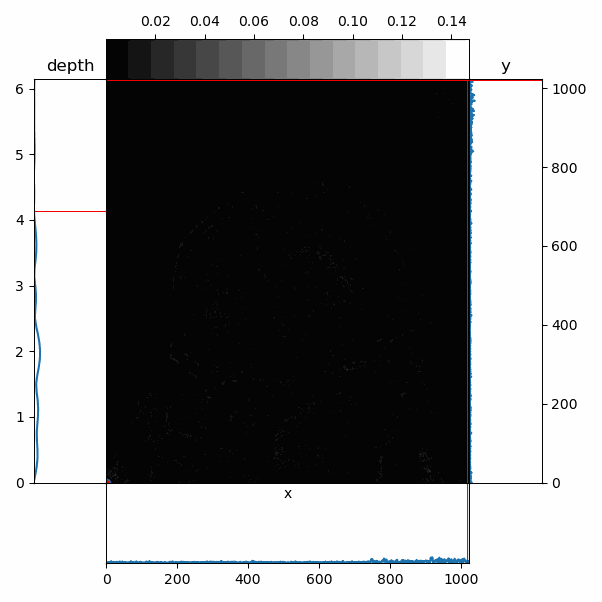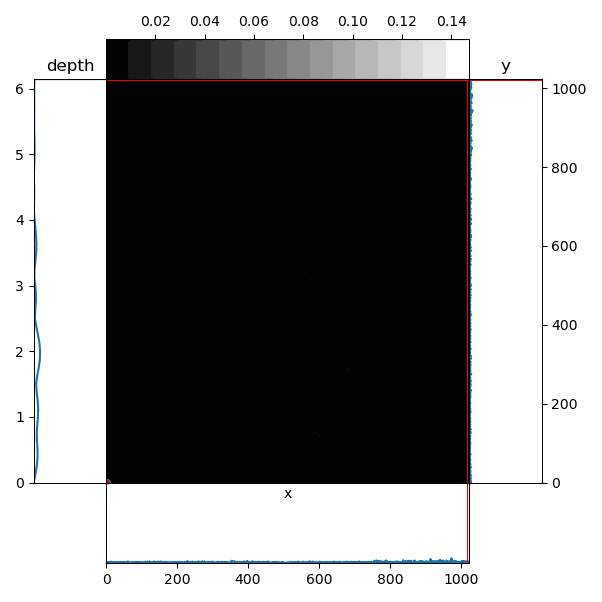
Brief example of basic PWS analysis to produce Sigma, Reflectance, Ld, and other images.¶
1# -*- coding: utf-8 -*-
2# Copyright 2018-2021 Nick Anthony, Backman Biophotonics Lab, Northwestern University
3#
4# This file is part of PWSpy.
5#
6# PWSpy is free software: you can redistribute it and/or modify
7# it under the terms of the GNU General Public License as published by
8# the Free Software Foundation, either version 3 of the License, or
9# (at your option) any later version.
10#
11# PWSpy is distributed in the hope that it will be useful,
12# but WITHOUT ANY WARRANTY; without even the implied warranty of
13# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
14# GNU General Public License for more details.
15#
16# You should have received a copy of the GNU General Public License
17# along with PWSpy. If not, see <https://www.gnu.org/licenses/>.
18
19"""
20This script saves pws analysis results to the test data. This must be run before many of the other examples will work.
21"""
22
23from pwspy import analysis
24from pwspy import dataTypes as pwsdt
25from examples import PWSExperimentPath
26
27settings = analysis.pws.PWSAnalysisSettings.loadDefaultSettings("Recommended")
28
29# Load our blank reference image which will be used for normalization.
30refAcq = pwsdt.Acquisition(PWSExperimentPath / 'Cell3')
31ref = refAcq.pws.toDataClass()
32
33anls = analysis.pws.PWSAnalysis(settings=settings, extraReflectance=None, ref=ref) # Create a new analysis for the given reference image and analysis settings. The "ExtraReflection" calibration is ignored in this case.
34
35acq = pwsdt.Acquisition(PWSExperimentPath / "Cell1") # Create an "Acquisition" object to handle operations for the data associated with a single acquisition
36cube = acq.pws.toDataClass() # Request that the PWS metadata object load the full data.
37results, warnings = anls.run(cube) # Run the pre-setup analysis on our data. Get the analysis results and potentially a list of warnings from the analysis.
38
39acq.pws.saveAnalysis(results, 'myAnalysis', overwrite=True) # Save our analysis results to file in the default location alongside the raw data under the `analyses` folder.
40
41loadedResults = acq.pws.loadAnalysis('myAnalysis') # This is just going to be a copy of `results`, loaded from file.
Use the compilation functionality to reduce analysis results to a table of values of average values within an ROI¶
1# Copyright 2018-2021 Nick Anthony, Backman Biophotonics Lab, Northwestern University
2#
3# This file is part of PWSpy.
4#
5# PWSpy is free software: you can redistribute it and/or modify
6# it under the terms of the GNU General Public License as published by
7# the Free Software Foundation, either version 3 of the License, or
8# (at your option) any later version.
9#
10# PWSpy is distributed in the hope that it will be useful,
11# but WITHOUT ANY WARRANTY; without even the implied warranty of
12# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
13# GNU General Public License for more details.
14#
15# You should have received a copy of the GNU General Public License
16# along with PWSpy. If not, see <https://www.gnu.org/licenses/>.
17
18"""
19This script loads analysis results from a list of PWS acquisitions and uses the "PWSRoiCompiler" class to get average output
20values within the ROIs for each acquisition. The compiled results are then placed into a Pandas dataframe.
21"""
22import pandas
23from pwspy.analysis.compilation import PWSRoiCompiler, PWSCompilerSettings
24from examples import PWSExperimentPath
25import pwspy.dataTypes as pwsdt
26
27tmpList = []
28compiler = PWSRoiCompiler(PWSCompilerSettings(reflectance=True, rms=True))
29
30listOfAcquisitions = [pwsdt.Acquisition(i) for i in PWSExperimentPath.glob("Cell[0-9]")]
31for acquisition in listOfAcquisitions:
32 for analysisName in acquisition.pws.getAnalyses():
33 analysisResults = acquisition.pws.loadAnalysis(analysisName)
34 for roiSpec in acquisition.getRois():
35 roiFile = acquisition.loadRoi(*roiSpec)
36 results, warnings = compiler.run(analysisResults, roiFile.getRoi())
37
38 if len(warnings) > 0:
39 print(warnings)
40
41 tmpList.append(dict(
42 acquisition=acquisition,
43 cellNumber=acquisition.getNumber(),
44 analysisResults=analysisResults,
45 rms=results.rms,
46 reflectance=results.reflectance,
47 roiNum=roiFile.number,
48 roiName=roiFile.name
49 ))
50
51dataFrame = pandas.DataFrame(tmpList)
52print(dataFrame)
Basic loading of ROI’s to extract data from specific regions¶
1# -*- coding: utf-8 -*-
2# Copyright 2018-2021 Nick Anthony, Backman Biophotonics Lab, Northwestern University
3#
4# This file is part of PWSpy.
5#
6# PWSpy is free software: you can redistribute it and/or modify
7# it under the terms of the GNU General Public License as published by
8# the Free Software Foundation, either version 3 of the License, or
9# (at your option) any later version.
10#
11# PWSpy is distributed in the hope that it will be useful,
12# but WITHOUT ANY WARRANTY; without even the implied warranty of
13# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
14# GNU General Public License for more details.
15#
16# You should have received a copy of the GNU General Public License
17# along with PWSpy. If not, see <https://www.gnu.org/licenses/>.
18
19"""
20Loop through all ROIs for all acquisitions in a directory and plot a histogram of the RMS values within the ROI.
21"""
22
23import pwspy.dataTypes as pwsdt
24import matplotlib.pyplot as plt
25import pathlib
26import numpy as np
27from examples import PWSExperimentPath
28plt.ion()
29
30workingDirectory = PWSExperimentPath # The folder that all your acquisitions are saved under.
31analysisName = 'script' # This will often be "p0"
32
33def plotHist(roi, rms):
34 """
35 This function takes an ROI
36 and a 2D RMS image and plots a histogram of the RMS values inside the ROI
37 """
38 # Check input values just to be safe.
39 assert isinstance(roi, pwsdt.Roi) # Make sure roiFile variable is actually an ROI
40 assert isinstance(rms, np.ndarray) # Make sure the RMS image is an numpy array
41 assert roi.mask.shape == rms.shape # Make sure the ROI and RMS arrays have the same dimensions.
42
43 vals = rms[roi.mask] # A 1D array of the values inside the ROI
44 plt.hist(vals) # Plot a histogram
45
46
47cellFolderIterator = pathlib.Path(workingDirectory).glob("Cell[0-9]") # An iterator for all folders that are below workingDirectory and match the "regex" pattern "Cell[0-9]"
48for folder in cellFolderIterator:
49 acq = pwsdt.Acquisition(folder) # An object handling the contents of a single "Cell{X}" folder
50
51 try:
52 anls = acq.pws.loadAnalysis(analysisName) # Load the analysis results from file.
53 except:
54 print(f"Analysis loading failed for {acq.filePath}")
55 continue # Skip to the next loop iteration
56
57 roiSpecs = acq.getRois() # A list of the names, numbers, and fileFormats of the ROIs in this acquisition
58
59 for name, number, fformat in roiSpecs: # Loop through every ROI.
60 roiFile = acq.loadRoi(name, number, fformat) # Load the ROI from file.
61 plotHist(roiFile.getRoi(), anls.rms) # Use the function defined above to plot a histogram
Using a hand-drawn ROI to generate a reference pseudo-measurement¶
1
2# -*- coding: utf-8 -*-
3# Copyright 2018-2021 Nick Anthony, Backman Biophotonics Lab, Northwestern University
4#
5# This file is part of PWSpy.
6#
7# PWSpy is free software: you can redistribute it and/or modify
8# it under the terms of the GNU General Public License as published by
9# the Free Software Foundation, either version 3 of the License, or
10# (at your option) any later version.
11#
12# PWSpy is distributed in the hope that it will be useful,
13# but WITHOUT ANY WARRANTY; without even the implied warranty of
14# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
15# GNU General Public License for more details.
16#
17# You should have received a copy of the GNU General Public License
18# along with PWSpy. If not, see <https://www.gnu.org/licenses/>.
19
20"""
21This script allows the user to select a region of an PwsCube. the spectra of this
22region is then averaged over the X and Y dimensions. This spectra is then saved
23as a reference dataTypes with the same initial dimensions.
24Can help to make a reference when you don't actually have one for some reason
25"""
26
27import pwspy.dataTypes as pwsdt
28import matplotlib.pyplot as plt
29import numpy as np
30from examples import PWSImagePath
31
32plt.ion()
33a = pwsdt.Acquisition(PWSImagePath).pws.toDataClass() # Load a measurement from file.
34
35roi = a.selectLassoROI() # Prompt the user for a hand-drawn ROI
36spec, std = a.getMeanSpectra(mask=roi) # Get the average spectra within the ROI
37newData = np.zeros(a.data.shape)
38newData[:, :, :] = spec[np.newaxis, np.newaxis, :] # Extend the averaged spectrum along the full dimensions of the original measurement.
39ref = pwsdt.PwsCube(newData, a.metadata) # Create a new synthetic measurement using the averaged spectrum
40plt.plot(a.wavelengths, spec)
41
Blurring data laterally to smooth a reference image.¶
1# -*- coding: utf-8 -*-
2# Copyright 2018-2021 Nick Anthony, Backman Biophotonics Lab, Northwestern University
3#
4# This file is part of PWSpy.
5#
6# PWSpy is free software: you can redistribute it and/or modify
7# it under the terms of the GNU General Public License as published by
8# the Free Software Foundation, either version 3 of the License, or
9# (at your option) any later version.
10#
11# PWSpy is distributed in the hope that it will be useful,
12# but WITHOUT ANY WARRANTY; without even the implied warranty of
13# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
14# GNU General Public License for more details.
15#
16# You should have received a copy of the GNU General Public License
17# along with PWSpy. If not, see <https://www.gnu.org/licenses/>.
18
19"""
20This script blurs an image cube in the xy direction. Allows you to turn an
21image of cells into something that can be used as a reference image, assuming
22most of the the FOV is glass. In reality you should just have a good reference image to use and not resort to something
23like this.
24"""
25
26import copy
27
28import matplotlib.pyplot as plt
29import pwspy.dataTypes as pwsdt
30from examples import PWSImagePath
31
32plt.ion()
33
34acq = pwsdt.Acquisition(PWSImagePath)
35a = acq.pws.toDataClass()
36
37a.correctCameraEffects() # Correct for dark counts and potentially for camera nonlinearity using metadata stored with the original measurement.
38a.normalizeByExposure() # Divide by exposure time to get data in units of `counts/ms`. This isn't strictly necessary in this case.
39
40mirror = copy.deepcopy(a)
41mirror.filterDust(10) # Apply a gaussian blurring with sigma=10 microns along the XY plane.
42
43a.plotMean() # Plot the mean reflectance of the original
44mirror.plotMean() # Plot the mean reflectance after filtering.
45a.normalizeByReference(mirror) # Normalize raw by reference
46a.plotMean() # Plot the measurement after normalization.
47plt.figure()
48plt.imshow(a.data.std(axis=2)) # Plot RMS after normalization.
Measuring Sigma using only a limited range of the OPD signal.¶
1
2# -*- coding: utf-8 -*-
3# Copyright 2018-2021 Nick Anthony, Backman Biophotonics Lab, Northwestern University
4#
5# This file is part of PWSpy.
6#
7# PWSpy is free software: you can redistribute it and/or modify
8# it under the terms of the GNU General Public License as published by
9# the Free Software Foundation, either version 3 of the License, or
10# (at your option) any later version.
11#
12# PWSpy is distributed in the hope that it will be useful,
13# but WITHOUT ANY WARRANTY; without even the implied warranty of
14# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
15# GNU General Public License for more details.
16#
17# You should have received a copy of the GNU General Public License
18# along with PWSpy. If not, see <https://www.gnu.org/licenses/>.
19
20"""
21This script is based on a matlab script written by Lusik Cherkezyan for NC.
22Nano uses this method to extract rms from phantom make from ChromEM cells embedded in resin.
23The phantom has a strong thin-film spectrum. This script is meant to filter out the thin film components
24of the fourier transfrom and extract RMS from what is left.
25"""
26from pwspy.dataTypes import CameraCorrection, Acquisition, Roi, PwsCube, KCube
27import matplotlib.pyplot as plt
28import scipy.signal as sps
29import os
30import numpy as np
31
32'''User Input'''
33path = r'2_7_2019 11.07'
34refName = 'Cell999' # This is an PwsCube of glass, used for normalization.
35cellNames = ['Cell1', 'Cell2'] # , 'Cell3', 'Cell4','Cell5']
36maskSuffix = 'resin'
37
38# identify the depth in um to which the OPD spectra need to be integrated
39integrationDepth = 2.0 ## in um
40isHannWindow = True # Should Hann windowing be applied to eliminate edge artifacts?
41subtractResinOpd = True
42resetResinMasks = False
43wvStart = 510 # start wavelength for poly subtraction
44wvEnd = 690 # end wavelength for poly subtraction
45sampleRI = 1.545 # The refractive index of the resin. This is taken from matlab code, I don't know if it's correct.
46orderPolyFit = 0
47wv_step = 2
48correction = CameraCorrection(2000, (0.977241216, 1.73E-06, 1.70E-11))
49
50'''************'''
51
52b, a = sps.butter(6, 0.1 * wv_step)
53opdIntegralEnd = integrationDepth * 2 * sampleRI # We need to convert from our desired depth into an opd value. There are some questions about having a 2 here but that's how it is in the matlab code so I'm keeping it.
54
55### load and save mirror or glass image cube
56ref = PwsCube.fromMetadata(Acquisition(os.path.join(path, refName)).pws)
57ref.correctCameraEffects(correction)
58ref.filterDust(6, pixelSize=1)
59ref.normalizeByExposure()
60
61if subtractResinOpd:
62 ### load and save reference empty resin image cube
63 fig, ax = plt.subplots()
64 resinOpds = {}
65 for cellName in cellNames:
66 resin = PwsCube.fromMetadata(Acquisition(os.path.join(path, cellName)).pws)
67 resin.correctCameraEffects(correction)
68 resin.normalizeByExposure()
69 resin /= ref
70 resin = KCube.fromPwsCube(resin)
71 if resetResinMasks:
72 [resin.metadata.acquisitionDirectory.deleteRoi(name, num) for name, num, fformat in resin.metadata.acquisitionDirectory.getRois() if name == maskSuffix]
73 if maskSuffix in [name for name, number, fformat in resin.metadata.acquisitionDirectory.getRois()]:
74 resinRoi = resin.metadata.acquisitionDirectory.loadRoi(maskSuffix, 1)
75 else:
76 print('Select a region containing only resin.')
77 resinRoi = resin.selectLassoROI()
78 resin.metadata.acquisitionDirectory.saveRoi(maskSuffix, 1, resinRoi)
79 resin.data -= resin.data.mean(axis=2)[:, :, np.newaxis]
80 opdResin, xvals = resin.getOpd(isHannWindow, indexOpdStop=None, mask=resinRoi.mask)
81 resinOpds[cellName] = opdResin
82 ax.plot(xvals, opdResin, label=cellName)
83 ax.vlines([opdIntegralEnd], ymin=opdResin.min(), ymax=opdResin.max())
84 ax.set_xlabel('OPD')
85 ax.set_ylabel("Amplitude")
86 ax.legend()
87 plt.pause(0.2)
88
89rmses = {} # Store the rms maps for later saving
90for cellName in cellNames:
91 cube = PwsCube.fromMetadata(Acquisition(os.path.join(path, cellName)).pws)
92 cube.correctCameraEffects(correction)
93 cube.normalizeByExposure()
94 cube /= ref
95 cube.data = sps.filtfilt(b, a, cube.data, axis=2)
96 cube = KCube.fromPwsCube(cube)
97
98 ## -- Polynomial Fit
99 print("Subtracting Polynomial")
100 polydata = cube.data.reshape((cube.data.shape[0] * cube.data.shape[1], cube.data.shape[2]))
101 polydata = np.rollaxis(polydata, 1) # Flatten the array to 2d and put the wavenumber axis first.
102 cubePoly = np.zeros(polydata.shape) # make an empty array to hold the fit values.
103 polydata = np.polyfit(cube.wavenumbers, polydata,
104 orderPolyFit) # At this point polydata goes from holding the cube data to holding the polynomial values for each pixel. still 2d.
105 for i in range(orderPolyFit + 1):
106 cubePoly += (np.array(cube.wavenumbers)[:, np.newaxis] ** i) * polydata[i,
107 :] # Populate cubePoly with the fit values.
108 cubePoly = np.moveaxis(cubePoly, 0, 1)
109 cubePoly = cubePoly.reshape(cube.data.shape) # reshape back to a cube.
110 # Remove the polynomial fit from filtered cubeCell.
111 cube.data = cube.data - cubePoly
112
113 rmsData = np.sqrt(np.mean(cube.data ** 2, axis=2)) #This can be compared to rmsOPDIntData, when the integralStopIdx is high and we don't do spectral subtraction they should be equivalent.
114
115 # Find the fft for each signal in the desired wavelength range
116 opdData, xvals = cube.getOpd(isHannWindow, None)
117
118 if subtractResinOpd:
119 opdData = opdData - resinOpds[cellName]
120
121 try:
122 integralStopIdx = np.where(xvals >= opdIntegralEnd)[0][0]
123 except IndexError: # If we get an index error here then our opdIntegralEnd is probably bigger than we can achieve. Just use the biggest value we have.
124 integralStopIdx = None
125 opdIntegralEnd = max(xvals)
126 print(f'Integrating to OPD {opdIntegralEnd}')
127
128 opdSquared = np.sum(opdData[:, :, :integralStopIdx] ** 2,axis=2) # Parseval's theorem tells us that this is equivalent to the sum of the squares of our original signal
129 opdSquared *= len(cube.wavenumbers) / opdData.shape[2] # If the original data and opd were of the same length then the above line would be correct. Since the fft has been upsampled. we need to normalize.
130 rmsOpdIntData = np.sqrt(opdSquared) # this should be equivalent to normal RMS if our stop index is high and resin subtraction is disabled.
131
132 cmap = plt.get_cmap('jet')
133 fig, axs = plt.subplots(1, 2, sharex=True, sharey=True)
134 im = axs[0].imshow(rmsData, cmap=cmap, clim=[np.percentile(rmsData, 0.5), np.percentile(rmsData, 99.5)])
135 fig.colorbar(im, ax=axs[0])
136 axs[0].set_title('RMS')
137 im = axs[1].imshow(rmsOpdIntData, cmap=cmap,
138 clim=[np.percentile(rmsOpdIntData, 0.5), np.percentile(rmsOpdIntData, 99.5)])
139 fig.colorbar(im, ax=axs[1])
140 axs[1].set_title(f'RMS from OPD below {opdIntegralEnd} after resin OPD subtraction')
141 fig.suptitle(cellName)
142 rmses[cellName] = rmsOpdIntData
143 plt.pause(0.2)
144plt.pause(0.5)
Generating new position lists to enable colocalized measurements on multiple systems.¶
1# Copyright 2018-2020 Nick Anthony, Backman Biophotonics Lab, Northwestern University
2#
3# This file is part of PWSpy.
4#
5# PWSpy is free software: you can redistribute it and/or modify
6# it under the terms of the GNU General Public License as published by
7# the Free Software Foundation, either version 3 of the License, or
8# (at your option) any later version.
9#
10# PWSpy is distributed in the hope that it will be useful,
11# but WITHOUT ANY WARRANTY; without even the implied warranty of
12# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
13# GNU General Public License for more details.
14#
15# You should have received a copy of the GNU General Public License
16# along with PWSpy. If not, see <https://www.gnu.org/licenses/>.
17from pwspy.utility.micromanager import PropertyMap
18
19from pwspy.utility.micromanager.positions import PositionList
20
21"""This example demonstrates how to use generate new cell positions from a set of positions after the sample has been picked up and likely shifted or rotated.
22This method relies on measuring a set (at least 3) of reference positions before and after moving the dish. You can then use these positions to generate an
23affine transform. This affine transform can then be applied to your original cell positions in order to generate a new set of positions for the same cells.
24In the case of a standard cell culture dish it is best to use the corners of the glass coverslip as your reference locations.
25
26@author: Nick Anthony
27"""
28
29if __name__ == '__main__':
30
31 import pathlib as pl
32 import matplotlib.pyplot as plt
33 from skimage.transform import AffineTransform
34 import numpy as np
35
36 plt.ion()
37 NCPath = pl.Path(r'Z:\Nick\NU-NC_EtOH fixation comparison\NC\Buccal cell')
38 NUPath = pl.Path(r'Z:\Nick\NU-NC_EtOH fixation comparison\NU\Buccal')
39
40 # Load the position list of the coverslip corners taken at the beginning of the experiment.
41 preTreatRefPositions = PositionList.fromNanoMatFile(NCPath / 'corners_list2.mat', 'TIXYDrive')
42 # Load the position list of the coverslip corners after placing the dish back on the microscope after treatment.
43 postTreatRefPositions = PositionList.fromPropertyMap(PropertyMap.loadFromFile(NUPath / 'corners.pos'))
44 # Generate an affine transform describing the difference between the two position lists.
45 transformMatrix = preTreatRefPositions.getAffineTransform(postTreatRefPositions)
46 # Load the positions of the cells we are measuring before the dish was removed.
47 preTreatCellPositions = PositionList.fromNanoMatFile(NCPath / 'cell_list.mat', 'TIXYDrive')
48 # Transform the cell positions to the new expected locations.
49 postTreatCellPositions = preTreatCellPositions.applyAffineTransform(transformMatrix)
50 # Save the new positions to a file that can be loaded by Micro-Manager.
51 postTreatCellPositions.toPropertyMap().saveToFile(NUPath / 'transformedPositions.pos')
52
53 # Plot the reference and cell positions before treatment
54 fig, ax = plt.subplots()
55 preTreatRefPositions.plot(fig, ax)
56 preTreatCellPositions.plot(fig, ax)
57
58 # Plot the reference and cell positions after treatment
59 fig2, ax2 = plt.subplots()
60 postTreatRefPositions.plot(fig2, ax2)
61 postTreatCellPositions.plot(fig2, ax2)
62
63 af = AffineTransform(np.vstack([transformMatrix, [0, 0, 1]]))
64 print(f"Scale: {af.scale}, Shear: {af.shear}, Rotation: {af.rotation / (2*np.pi) * 360}, Translation: {af.translation}")